Train spatialMaxent models
We will now train two models for each species in the Canada dataset. One with a random 5-fold cross-validation and the Maxent default settings (default modeling approach) and one with spatial cross-validation and forward-variable-selection, forward-feature-selection and regularization multiplier tuning (spatial modeling approach). Both models are trained and evaluated on the exact same data and we will compare their performance in the next section.
We will need to create 21 models for each species, with all possible combinations of test and training data to get a comprehensive picture, which modelling approach performs best. As we are comparing two modeling approaches for 20 species we will calculate a total of 840 models, if you are not having sufficient free storage on your PC (~ 33 GB) you can delete the prediction rasters once all metrics are calculated.
spatialMaxent GUI
Watch the video below to see how to train the model for the species can01 and the first combination of training and test data can01_01 in the spatialMaxent GUI.
spatialMaxent from the command line
As we are training quite a few models here: 21 models per species for 20 species for 2 different modeling approaches we have a total of 840 models to train, to do this we will not rely on the GUI but use a R script to call spatialMaxent from the command line. For the default modeling approach, we will set the parameters fvs, ffs and tuneRMto false and chose crossvalidate as replicate type. We are also projecting the models with the raster layers to the whole study area. In the script below the number of threads is set to 4. Adjust this number to the capacities of your PC.
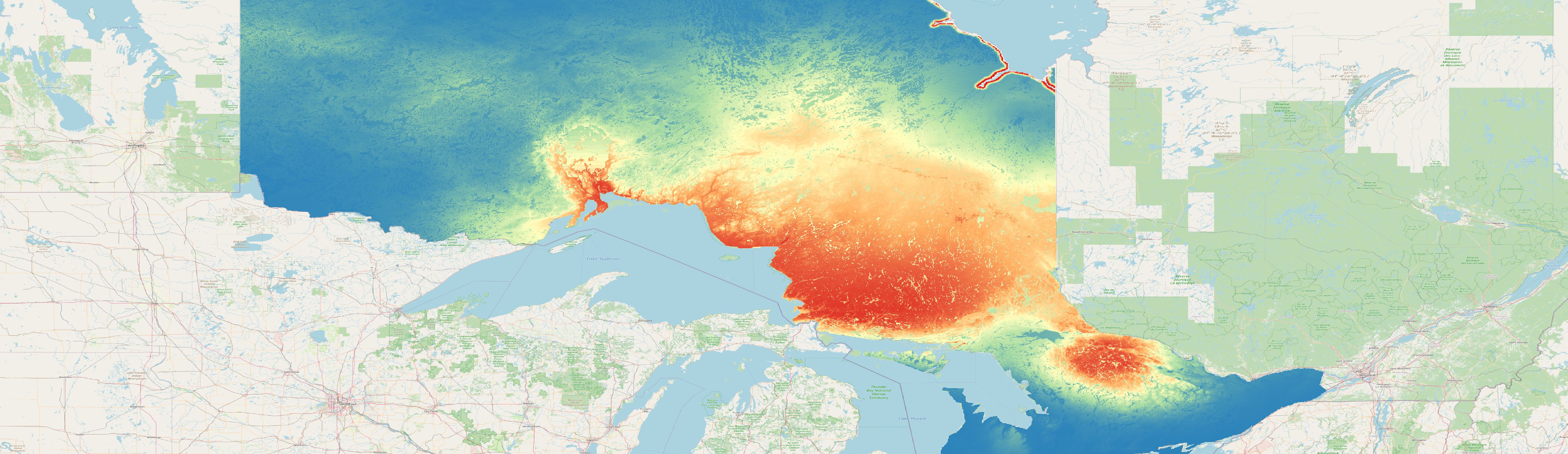
Prediction - spatial model
Exemplary prediction for the species can01and the fold can01_01 created with the spatial modeling approach.
Full screen version of the map
Prediction - default model
Exemplary prediction for the species can01and the fold can01_01 created with the default modeling approach.